I am trying to scrape information about orbits for asteroids in R. I have tried rvest and selectorgadget, however the website is dynamic. The website is: https://ssd.jpl.nasa.gov/tools/sbdb_lookup.html#/?sstr=2006%20WP1
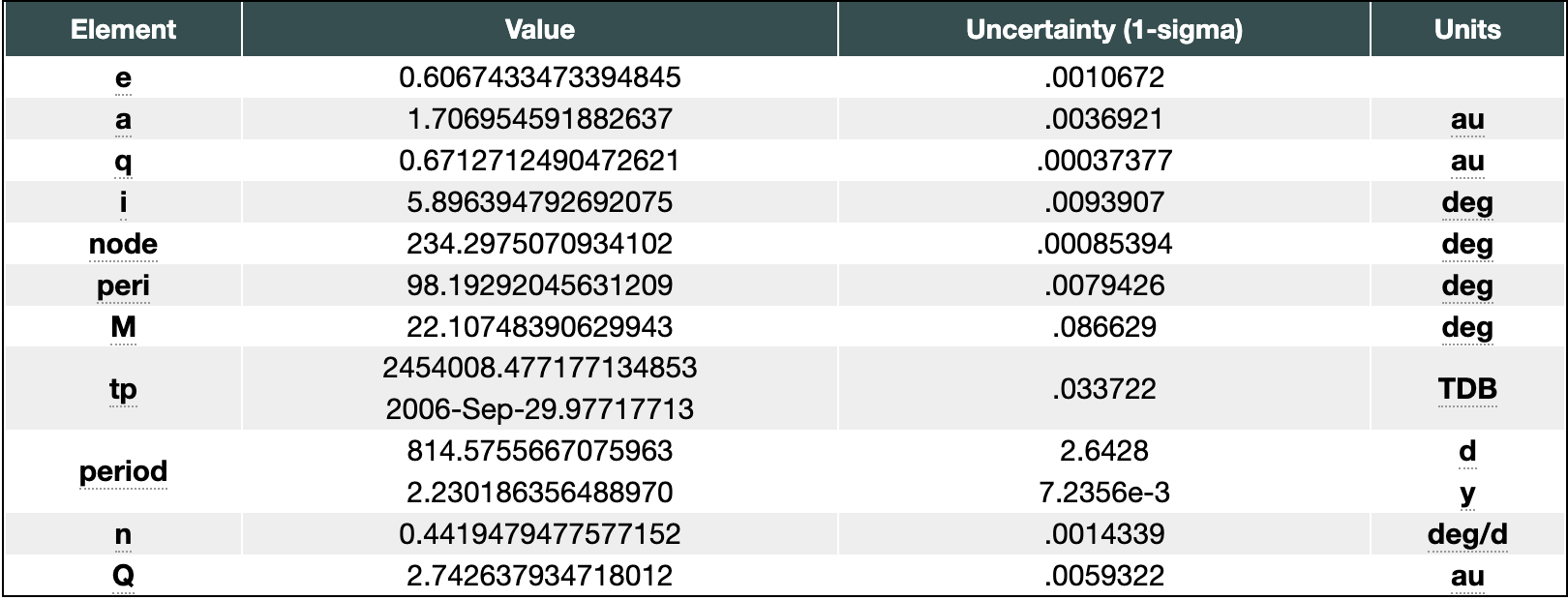
I am wanting to get the data for the Osculating Orbital Elements under the Orbit Parameters drop down shown here:
I am not very familiar with html or json, so I am looking for help downloading this table into R.

 Question posted in
Question posted in 

2
Answers
Use their API.
As an example of how to use the API, inspecting the page we can see the parameters used and recreate them (although I have changed it to use the direct identifier rather than a search string because it is quicker):
Now retrieve the data:
To complete, using rvest
read_html_live:Output :