I am working with the R programming language.
For the following website : https://covid.cdc.gov/covid-data-tracker/ – I am trying to get all versions of this website that are available on WayBackMachine (along with the month and time). The final result should look something like this:
date links time
1 Jan-01-2023 https://web.archive.org/web/20230101000547/https://covid.cdc.gov/covid-data-tracker/ 00:05:47
2 Jan-01-2023 https://web.archive.org/web/20230101000557/https://covid.cdc.gov/covid-data-tracker/ 00:05-57
Here is what I tried so far:
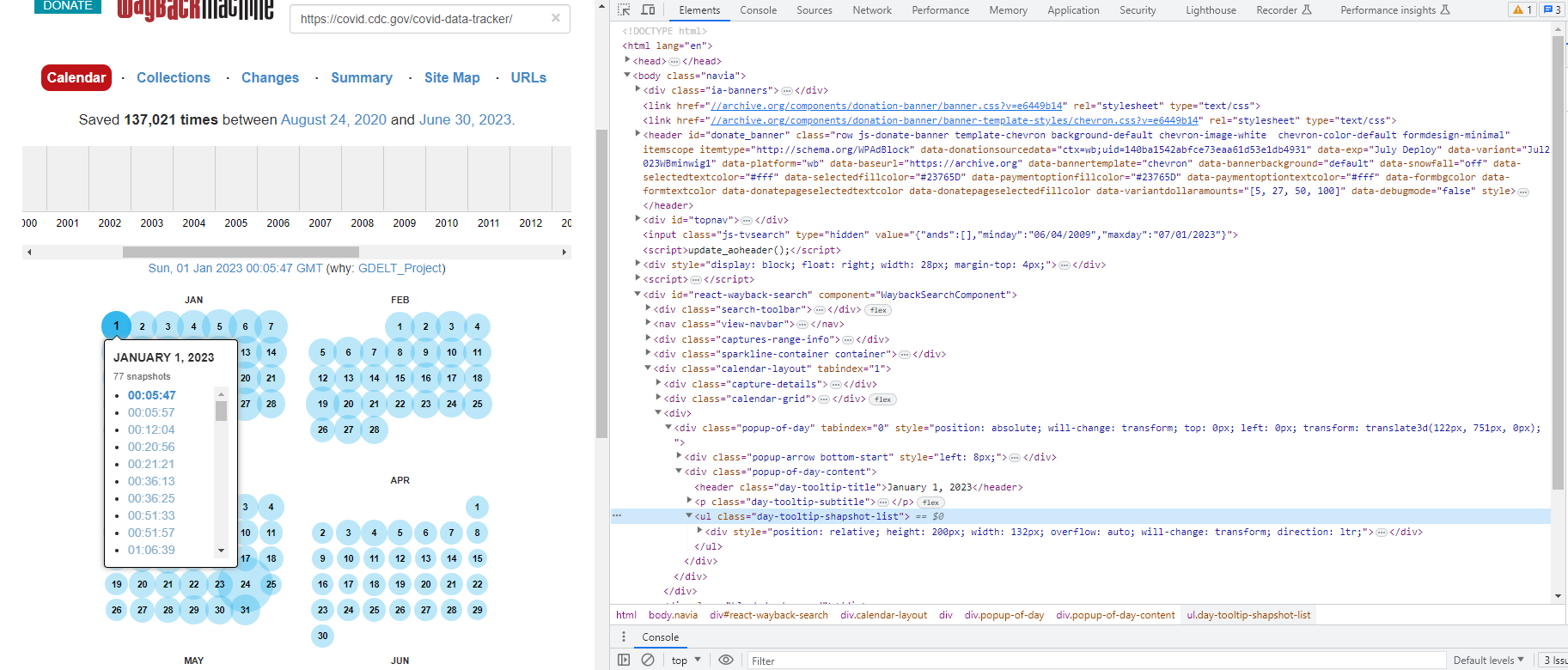
Step 1: First, I inspected the HTML source code (within "elements") and copied/pasted it into a notepad file:
Step 2: Then, I imported this into R and parsed the resulting html for the link structure:
file <- "cdc.txt"
text <- readLines(file)
html <- paste(text, collapse = "n")
pattern1 <- '/web/\d+/https://covid\.cdc\.gov/covid-data-tracker/[^"]+'
links <- regmatches(html, gregexpr(pattern1, html))[[1]]
But this is not working :
> links
character(0)
Can someone please show me if there is an easier way to do this?
Thanks!
Note:
-
I am trying to learn how to do this in general (i.e. for any websites on WayBackMachine – the Covid Data Tracker is just an placeholder example for this question)
-
I realize that there might be much more efficient ways to do this – I open to learning about different approaches!




2
Answers
This is really two questions. The html is generated client side rather than server side. This is why you cannot just request the html from R to get what you need, and end up copying and pasting from Developer Tools. You can automate this by using RSelenium. The docs are extensive so I won’t cover that in the answer.
You should also use a parser like rvest to parse the html, rather than regular expressions. In this case, to get the output you want, that would look something like:
Archive.org provides Wayback CDX API for looking up captures, it returns timestamps along with original urls in tabular form or JSON. Such queries can be made with
read.table()alone, links to specific captures can then be constructed fromtimestampandoriginalcolumns and base URL.To make it a bit more convenient to work with, we can customize API request with
httr/httr2, for example, and pass the response throughreadr/dplyr/lubridatepipeline:Created on 2023-07-02 with reprex v2.0.2
There are also Archive.org client libraries for R, e.g.
https://github.com/liserman/archiveRetriever & https://hrbrmstr.github.io/wayback/ , though the query interface for the first is bit odd, and the other is currently not available through CRAN.