I have a simple loop that just sums the second row of a numpy array. In numba I need only do:
from numba import njit
@njit('float64(float64[:, ::1])', fastmath=True)
def fast_sum_nb(array_2d):
s = 0.0
for i in range(array_2d.shape[1]):
s += array_2d[1, i]
return s
If I time the code I get:
In [3]: import numpy as np
In [4]: A = np.random.rand(2, 1000)
In [5]: %timeit fast_sum_nb(A)
305 ns ± 7.81 ns per loop (mean ± std. dev. of 7 runs, 1,000,000 loops each)
To do the same thing in cython I need first make setup.py which has:
from setuptools import setup
from Cython.Build import cythonize
from setuptools.extension import Extension
ext_modules = [
Extension(
'test_sum',
language='c',
sources=['test.pyx'], # list of source files
extra_compile_args=['-Ofast', '-march=native'], # example extra compiler arguments
)
]
setup(
name = "test module",
ext_modules = cythonize(ext_modules, compiler_directives={'language_level' : "3"})
)
I have the highest possible compilation options. The cython summation code is then:
#cython: language_level=3
from cython cimport boundscheck
from cython cimport wraparound
@boundscheck(False)
@wraparound(False)
def fast_sum(double[:, ::1] arr):
cdef int i=0
cdef double s=0.0
for i in range(arr.shape[1]):
s += arr[1, i]
return s
I compile it with:
python setup.py build_ext --inplace
If I now time this I get:
In [2]: import numpy as np
In [3]: A = np.random.rand(2, 1000)
In [4]: %timeit fast_sum(A)
564 ns ± 1.25 ns per loop (mean ± std. dev. of 7 runs, 1,000,000 loops each)
Why is the cython version so much slower?
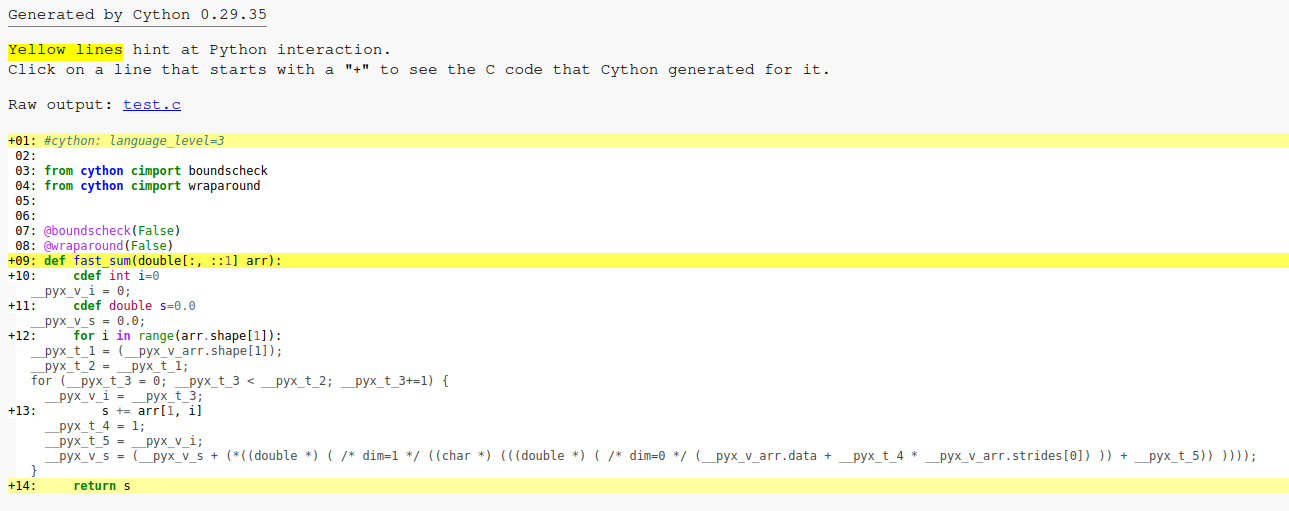
The annotated C code from cython looks like this:
The assembly produced by numba seems to be:
vaddpd -96(%rsi,%rdx,8), %ymm0, %ymm0
vaddpd -64(%rsi,%rdx,8), %ymm1, %ymm1
vaddpd -32(%rsi,%rdx,8), %ymm2, %ymm2
vaddpd (%rsi,%rdx,8), %ymm3, %ymm3
addq $16, %rdx
cmpq %rdx, %rcx
jne .LBB0_5
vaddpd %ymm0, %ymm1, %ymm0
vaddpd %ymm0, %ymm2, %ymm0
vaddpd %ymm0, %ymm3, %ymm0
vextractf128 $1, %ymm0, %xmm1
vaddpd %xmm1, %xmm0, %xmm0
vpermilpd $1, %xmm0, %xmm1
vaddsd %xmm1, %xmm0, %xmm0
I don’t know how to get the assembly for the cython code. The C file it produces is huge and the .so file disassembles to a large file as well.
This speed difference persists (in fact it increases) if I increase the number of columns in the 2d array so it doesn’t seem to be a calling overhead issue.
I am using Cython version 0.29.35 and numba version 0.57.0 on Ubuntu.




2
Answers
It looks like
is just producing better-optimized code than
There are three things that conspire to give Cython worse performance in this benchmark.
Cython overhead (minor)
The loop Cython generates (major)
Which compiler is being used (major)
Overhead (reason 1)
Cython apparently has some extra overhead. You can observe this by passing in an array of shape (1,0).
The Numba function is still much faster.
This isn’t too surprising, given that Cython is the more general tool, and it tries to be extra careful with inputs, error handling, etc., even when it’s overkill.
Unless you’re calling this function a lot with very small inputs, this shouldn’t matter much.
Loop Unrolling (reasons 2 & 3 together)
Based on the disassembly in the updated question (copied here), Numba + LLVM are creating a nicely unrolled loop.
Notice how it’s using YMM0..YMM3, not just one vector register.
In contrast, here’s core decompiled loop from Cython when using gcc. There’s no unrolling here.
The decompilation of the clang output is similar, but happens to perform even worse (see the benchmark results below). For some reason, clang doesn’t want to unroll Cython’s loop.
Making Cython Fast
It can sometimes be tricky to make Cython generate ultra-fast code, but fortunately there’s another option: use Cython only for glue between Python and C.
Try putting this in your
.pyxfile:and then create an
impl.hfile with the following contents:On my machine, with a (2,1000) input array, here are the
timeitruntimes:Observations:
Numba has a little lower overhead than Cython.
Cython + Clang performs exceptionally poorly on this benchmark.
…but a thin Cython wrapper + hand written C code + Clang is almost as good as Numba.
GCC doesn’t seem as sensitive to the code Cython generates. We get the same speed with Cython’s code and hand written code.
Here’s the most important assembly snippet from the
clang-compiled version offast_sum_c.Unsurprisingly, it’s very similar to what Numba produced (since they both use LLVM as their backend):
Notes
Compiler options to encourage more loop unrolling didn’t help. In the "Making Cython Fast", I tried adding various pragmas and compiler flags to gcc to encourage it to do some unrolling; nothing helped. Additionally, clang never had trouble with my hand written C code, but never wanted to unroll the Cython-generated loop.
objdump -d test_sum.*.soproduces a disassembly. Looking for thevaddpdinstructions helps with locating the loop of interest.Ghidra can be used to decompile the code. This helps a little with understanding it. Compiling the extension module with
-gand-gdwarf-4makes it so Ghidra’s DWARF decoding works, injecting a bit more metadata in the decompilation.For these tests, I used
clang14.0.0 andgcc11.3.0.TL;DR: this answer adds additional details on top of the good answer of @MrFooz so to understand why the Cython code is slow with both Clang and GCC. The performance issue is coming from a combination of 3 missed optimizations : one from Clang, one from GCC and one from Cython…
Under the hood
First of all, Cython generates a C code with a stride not known at compile time. This is a problem since auto-vectorized of compilers cannot easily vectorize the code using SIMD instruction as the array could theoretically not be contiguous (even though it will always be in practice). As a result, the Clang auto-vectorizer fail to optimize the loop (both auto-vectorization and unrolling). The GCC optimizer is a bit more clever : it generate a specialized vectorized code for a stride of 1 (ie. contiguous arrays). Here is the generated Cython code:
Note the
__pyx_v_arr.strides[0]which is not replaced by 1 at compile time while Cython is supposed to know the array is contiguous. There is a workaround for this Cython missed-optimization : using 1D memory-views.Here is the modified Cython code:
This code is unfortunately not faster because of two underlying compiler issues…
GCC does not unroll such a loop yet by default. This is a well-known long-standing missed optimization. In fact, there is even an open issue for this specific C code. Using the compilation flags
-funroll-loops -fvariable-expansion-in-unrollerhelps to improve the resulting performance though the generated code is still not perfect.When it comes to Clang, this is another missed optimization preventing the code to be fast. GCC and Clang had several open issues in the past related to missed auto-vectorization when using types of different size in loop to be vectorized (and even with signed/unsigned for GCC). To fix this issue, you should use 64-bit integers when using double-precision arrays. Here is the final modified Cython code:
Note that Numba use 64-bit integers by default (eg. for loop iterators and indices) and Numba use LLVM-Lite (based on LLVM, like Clang) so such a problem does not happen here.
Benchmark
Here are performance results on my machine with a i5-9600KF processor, GCC 12.2.0, Clang 14.0.6 and Python 3.11:
The very small overhead between Numba and Cython+Clang is due to different startup overheads. Generally, such a small time should not be an issue since one should not call Cython/Numba functions from CPython a lot of time. In such a pathological situation, the caller function should also be modified to use Cython/Numba. When this is not possible nether Numba nor Cython will produce a fast code, so lower-level C extension should be preferred instead.