I’m currently working my way through converting some C++ code from a paper to Python (a language I’m far more familiar with). I’ve gotten all the values to this point in the code to match (I managed to get the C++ repo from github to compile and can debug in Visual Studio to compare to my outputs in Python).
I seem to have hit a bit of a stumbling block with the way the dgesvd call is being made to lapack in the C++ code, as I’m not too sure how it’s being handled behind the scenes and how I can mimic the behaviour in Python.
The C++ code calls the lapack dgesvd as follows:
// Compute the SVD of K0
char save[]="S", nosave[]="N";
int nosavedim=1;
int info, lwork;
int cols_s = 316*dofn3_s;
lwork = 5*n3; // Change
work = (double *) malloc(lwork * sizeof(double));
int U0size;
Sigma_size = dofn3_f;
U0size = dofn3_f * dofn3_f;
Sigma = (double *)malloc(Sigma_size * sizeof(double));
U0 = (double *)malloc(U0size * sizeof(double));
VT0 = NULL;
dgesvd_(save, nosave, &dofn3_f, &cols_s, K0, &dofn3_f, Sigma, U0,
&dofn3_f, VT0, &nosavedim, work, &lwork, &info);
So, the variables going into the dgesvd call from C++ are:
| Variable | Value |
|---|---|
| save | "S" |
| nosave | "N" |
| dofn3_f | 27 |
| cols_s | 8532 |
| K0 | 1D array of shape (230364,) |
| dofn3_f | 27 |
| Sigma | Return from SVD |
| U0 | Return from SVD |
| dofn3_f | 27 |
| VT0 | NULL (don’t return from SVD) |
| nosavedim | 1 |
| work | 1809 |
| lwork | 135 |
| info | Return from SVD |
The C++ code gets back:
| Variable | Value |
|---|---|
| U0 | 1D array of shape (729,) |
| Sigma | 1D array of shape (27,) |
From what I can tell from the lapack doco this essentially means it’s asking dgesvd for the first min(m,n) columns of U (the left singular vectors) to be returned in the array U0 and no output for VT0 (which matches what’s returned).
What I don’t quite understand, though, is that the input array is 1D, yet the input matrix is supposed to be an M-by-N matrix. I’m assuming that the lapack function is converting it behind the scenes (or operating on it in the correct order based on the inputs)?
Looking at the low level Scipy wrapper for the lapack dgesvd and the Scipy svd itself and testing both, they both require the input array to be of shape (m, n).
I’ve tried calling the low level Scipy wrapper as follows:
l_work = np.int32(5 * self.n3)
U0, Sigma, V0, info = linalg.lapack.dgesvd(k0, full_matrices=0, lwork=l_work)
Which returns an error ** On entry to DGESVD parameter number 13 had an illegal value.
If I remove the lwork kwarg it doesn’t error, but returns:
| Variable | Value |
|---|---|
| U0 | 1D array of shape (23064,) |
| Sigma | 1D array of shape (1,) |
I’ve tried reshaping the input array to be of shape (27, 8532) by using np.reshape((27, 8532)) and using both the low level wrapper and the standard Scipy implementation, this returns:
| Variable | Value |
|---|---|
| U0 | 1D array of shape (27,27) |
| Sigma | 1D array of shape (27,) |
Which, if I reshape the U0 array, gives me the same shapes as the C++ gets back from lapack, but my values are all quite different to those from the C++ results.
Is this simply a matter of reshaping the arrays in a different order somehow? And if so, how would I need to do this to ensure I get the same values back from the SVD calc as the C++ code?
EDIT
To provide some further context, if I run:
A = K0.reshape((27, 8532))
U0, Sigma, V0, info = scipy.linalg.svd(A)
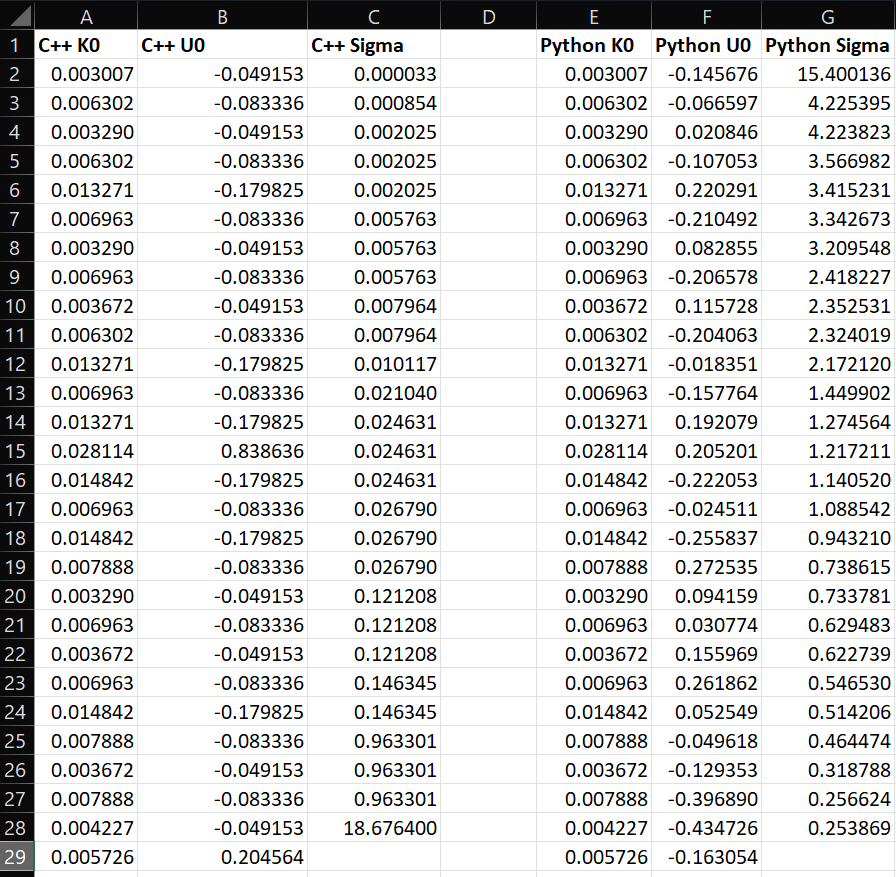
Here’s a screenshot of a comparison of the values I’m seeing from the C++ code and my Python version. You can see the input array K0 matches, but the values returned from the SVD calcs are different. The K0 array being parsed into the C++ lapack call is a 1D array, so it’s obviously converting it to a 2D array of shape (m, n) somehow behind the scenes – this is what I need to be able to mimic in Python to ensure I get the same answer out of the SVD.




2
Answers
Right, so I've been down a bit of a rabbit hole today, but think I've finally got this figured out.
If anyone comes searching for this in the future, turns out the array being parsed to the lapack dgesvd function in the C++ code is column-major, so to get the same values in Python I needed to use the following to convert the 1D column-major array to a numpy array to use in
scipy.linalg.svd, then flatten and reverse to convert back to give the same result as the C++ code:You should consider using
scipy.linalg.svdinstead of calling the underlying LAPACK routine.The code you pasted seems to only care about
UandSfrom the svd so getting the result you want might be as simple as:with SciPy handling all the underlying memory shapes for you. Here if
K0is (M,N) thenUwill be (M,M) which in your case should be (27,27) and have exactly 729 elements just like the C code.Calling the underlying LAPACK and BLAS code from python is often not needed as
scipyhas made wrappers for all the common routines.